Handling Missing Data - p.10 Data Analysis with Python and Pandas Tutorial
Welcome to Part 10 of our Data Analysis with Python and Pandas tutorial. In this part, we're going to be talking about missing or not available data. We have a few options when considering the existence of missing data.
- Ignore it - Just leave it there
- Delete it - Remove all cases. Remove from data entirely. This means forfeiting the entire row of data.
- Fill forward or backwards - This means taking the prior or following value and just filling it in.
- Replace it with something static - For example, replacing all NaN data with -9999.
Each of these options has their own merits for a variety of reasons. Ignoring it requires no more work on our end. You may choose to ignore missing data for legal reasons, or maybe to retain the utmost integrity of the data. Missing data might also be very important data. For example, maybe part of your analysis is investigating signal drops from a server. In this case, maybe the missing data is super important to keep in the set.
Next, we have delete it. You have another two choices at this point. You can either delete rows if they contain any amount of NaN data, or you can delete the row if it is completely NaN data. Usually a row that is full of NaN data comes from a calculation you performed on the dataset, and no data is really missing, it's just simply not available given your formula. In most cases, you would at least want to drop all rows that are completely NaN, and in many cases you would like to just drop rows that have any NaN data. How might we go about doing this? We'll start with the following script (notice the resampling is being done now by adding a new column to the HPI_data dataframe.):
import Quandl
import pandas as pd
import pickle
import matplotlib.pyplot as plt
from matplotlib import style
style.use('fivethirtyeight')
# Not necessary, I just do this so I do not show my API key.
api_key = open('quandlapikey.txt','r').read()
def state_list():
fiddy_states = pd.read_html('https://simple.wikipedia.org/wiki/List_of_U.S._states')
return fiddy_states[0][0][1:]
def grab_initial_state_data():
states = state_list()
main_df = pd.DataFrame()
for abbv in states:
query = "FMAC/HPI_"+str(abbv)
df = Quandl.get(query, authtoken=api_key)
print(query)
df[abbv] = (df[abbv]-df[abbv][0]) / df[abbv][0] * 100.0
print(df.head())
if main_df.empty:
main_df = df
else:
main_df = main_df.join(df)
pickle_out = open('fiddy_states3.pickle','wb')
pickle.dump(main_df, pickle_out)
pickle_out.close()
def HPI_Benchmark():
df = Quandl.get("FMAC/HPI_USA", authtoken=api_key)
df["United States"] = (df["United States"]-df["United States"][0]) / df["United States"][0] * 100.0
return df
##fig = plt.figure()
##ax1 = plt.subplot2grid((1,1), (0,0))
HPI_data = pd.read_pickle('fiddy_states3.pickle')
HPI_data['TX1yr'] = HPI_data['TX'].resample('A')
print(HPI_data[['TX','TX1yr']])
##HPI_data['TX'].plot(ax=ax1)
##HPI_data['TX1yr'].plot(color='k',ax=ax1)
##
##plt.legend().remove()
##plt.show()
We're commenting out the graphing stuff for now, but we'll revisit in a moment.
Output:
TX TX1yr Date 1975-01-31 0.000000 NaN 1975-02-28 1.291954 NaN 1975-03-31 3.348154 NaN 1975-04-30 6.097700 NaN 1975-05-31 6.887769 NaN 1975-06-30 5.566434 NaN 1975-07-31 4.710613 NaN 1975-08-31 4.612650 NaN 1975-09-30 4.831876 NaN 1975-10-31 5.192504 NaN 1975-11-30 5.832832 NaN 1975-12-31 6.336776 4.559105 1976-01-31 6.576975 NaN 1976-02-29 7.364782 NaN 1976-03-31 9.579950 NaN 1976-04-30 12.867197 NaN 1976-05-31 14.018165 NaN 1976-06-30 12.938501 NaN 1976-07-31 12.397848 NaN 1976-08-31 12.388581 NaN 1976-09-30 12.638779 NaN 1976-10-31 13.341849 NaN 1976-11-30 14.336404 NaN 1976-12-31 15.000798 11.954152 1977-01-31 15.555243 NaN 1977-02-28 16.921638 NaN 1977-03-31 20.118106 NaN 1977-04-30 25.186161 NaN 1977-05-31 26.260529 NaN 1977-06-30 23.430347 NaN ... ... ... 2011-01-31 280.574891 NaN 2011-02-28 281.202150 NaN 2011-03-31 282.772390 NaN 2011-04-30 284.374537 NaN 2011-05-31 286.518910 NaN 2011-06-30 288.665880 NaN 2011-07-31 288.232992 NaN 2011-08-31 285.507223 NaN 2011-09-30 283.408865 NaN 2011-10-31 282.348926 NaN 2011-11-30 282.026481 NaN 2011-12-31 282.384836 284.001507 2012-01-31 283.248573 NaN 2012-02-29 285.790368 NaN 2012-03-31 289.946517 NaN 2012-04-30 294.803887 NaN 2012-05-31 299.670256 NaN 2012-06-30 303.575682 NaN 2012-07-31 305.478743 NaN 2012-08-31 305.452329 NaN 2012-09-30 305.446084 NaN 2012-10-31 306.424497 NaN 2012-11-30 307.557154 NaN 2012-12-31 308.404771 299.649905 2013-01-31 309.503169 NaN 2013-02-28 311.581691 NaN 2013-03-31 315.642943 NaN 2013-04-30 321.662612 NaN 2013-05-31 328.279935 NaN 2013-06-30 333.565899 NaN [462 rows x 2 columns]
We have lots of NaN data. If we uncomment all the graphing code, what happens? Turns out, we don't get the graph that contains NaN data! This is a bummer, so first we're thinking, okay, let's drop all the rows that have any NaN data. This is just for tutorial purposes. In this example, that'd be a pretty bad idea to do. Instead, you would want to do what we originally did, which was create a new dataframe for the resampled data. This doesn't mean that's what you'd always do, but, in this case, you would. Anyway, let's drop all rows that contain any na data. That's as simple as:
HPI_data.dropna(inplace=True) print(HPI_data[['TX','TX1yr']])
TX TX1yr Date 1975-12-31 6.336776 4.559105 1976-12-31 15.000798 11.954152 1977-12-31 30.434104 23.518179 1978-12-31 51.029953 41.978042 1979-12-31 75.975953 64.700665 1980-12-31 89.979964 85.147662 1981-12-31 108.121926 99.016599 1982-12-31 118.210559 114.589927 1983-12-31 127.233791 122.676432 1984-12-31 133.599958 131.033359 1985-12-31 132.576673 133.847016 1986-12-31 126.581048 131.627647 1987-12-31 109.829893 119.373827 1988-12-31 104.602726 107.930502 1989-12-31 108.485926 107.311348 1990-12-31 109.082279 108.727174 1991-12-31 114.471725 113.142303 1992-12-31 121.427564 119.650162 1993-12-31 129.817931 127.009907 1994-12-31 135.119413 134.279735 1995-12-31 141.774551 139.197583 1996-12-31 146.991204 145.786792 1997-12-31 155.855049 152.109010 1998-12-31 170.625043 164.595301 1999-12-31 188.404171 181.149544 2000-12-31 206.579848 199.952853 2001-12-31 217.747701 215.692648 2002-12-31 230.161877 226.962219 2003-12-31 236.946005 235.459053 2004-12-31 248.031552 245.225988 2005-12-31 267.728910 260.589093 2006-12-31 288.009470 281.876293 2007-12-31 296.154296 298.094138 2008-12-31 288.081223 296.999508 2009-12-31 291.665787 292.160280 2010-12-31 281.678911 291.357967 2011-12-31 282.384836 284.001507 2012-12-31 308.404771 299.649905
No more rows with missing data!
Now, we can graph it:
fig = plt.figure()
ax1 = plt.subplot2grid((1,1), (0,0))
HPI_data = pd.read_pickle('fiddy_states3.pickle')
HPI_data['TX1yr'] = HPI_data['TX'].resample('A')
HPI_data.dropna(inplace=True)
print(HPI_data[['TX','TX1yr']])
HPI_data['TX'].plot(ax=ax1)
HPI_data['TX1yr'].plot(color='k',ax=ax1)
plt.legend().remove()
plt.show()
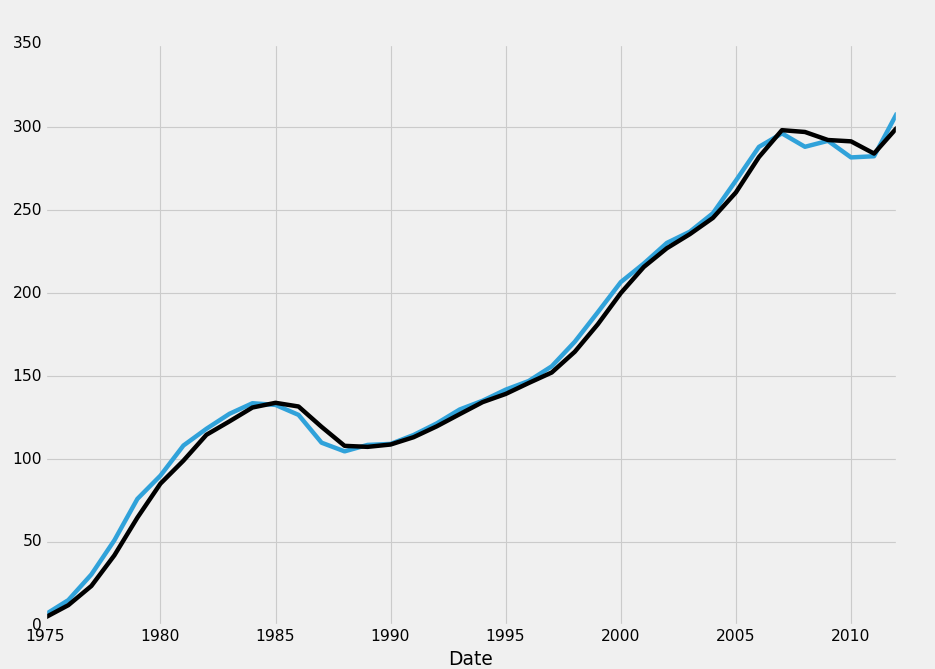
Okay, great. Now just for tutorial purposes, how might we write code that only deletes rows if the full row is NaN?
HPI_data.dropna(how='all',inplace=True)
For the "how" parameter, you can choose between all or any. All requires all data in the row to be NaN for you to delete it. You can also choose "any" and then set a threshold. This threshold will require that many non-NA values to accept the row. See the Pandas documentation for dropna for more information.
Alright, so that's dropna, next we have filling it. With filling, we have two major options again, which is whether to fill forward, backwards. The other option is to just replace the data, but we called that a separate choice. It just so happens that the same function is used to do it, fillna().
Modifying our original code block, with the main change of:
HPI_data.fillna(method='ffill',inplace=True)
Giving us:
fig = plt.figure()
ax1 = plt.subplot2grid((1,1), (0,0))
HPI_data = pd.read_pickle('fiddy_states3.pickle')
HPI_data['TX1yr'] = HPI_data['TX'].resample('A')
HPI_data.fillna(method='ffill',inplace=True)
HPI_data.dropna(inplace=True)
print(HPI_data[['TX','TX1yr']])
HPI_data['TX'].plot(ax=ax1)
HPI_data['TX1yr'].plot(color='k',ax=ax1)
plt.legend().remove()
plt.show()
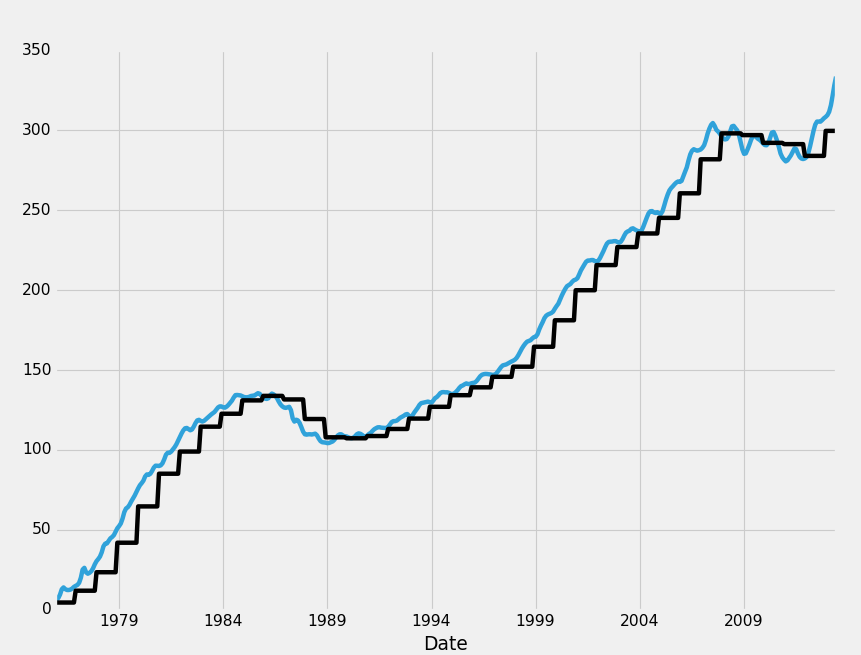
What ffill, or "forward fill" does for is is fills data forward into missing data. Think of it like a sweeping motion where you take data from earlier, sweeping it forward to the missing data. Any case of missing data will be filled witht he most recent non-missing data. Bfill, or backfilling is the opposite:
HPI_data.fillna(method='bfill',inplace=True)
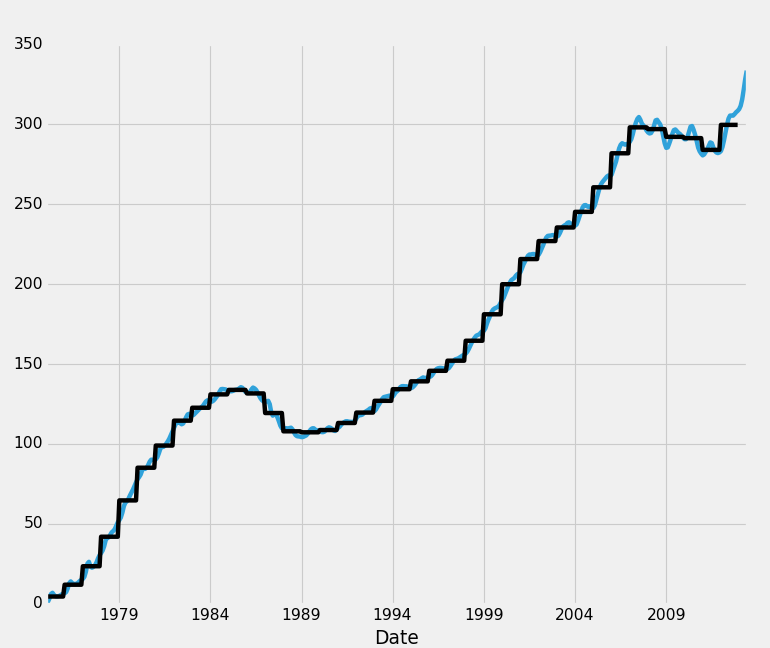
This takes data from the future, and sweeps it backwards to fill in holes.
Now, for the last method, replacing the data. NaN data is relatively worthless data, but it can taint the rest of our data. Take, for example, machine learning, where each row is a feature set and each column is a feature. Data has a very high value to us, and if we have a large amount of NaN data, forfeiting all of the data is a huge bummer. For this reason, you may actually use a replace instead. With most machine learning classifiers, extreme outliers are often ignored in the end as their own data point. Because of this, what many people will do is take any NaN data, and replace it with a value of, say, -99999. This is because, after pre-processing the data, you generally want to convert all features to a range of -1 to positive 1. A data point that is -99999 is a clear and obvious outlier to almost any classifier. NaN data, however, simply cannot be handled at all! Thus, we can replace data, by doing something like the following:
HPI_data.fillna(value=-99999,inplace=True)
Now, in our case, this is an absolutely useless move, but it does have its place in certain forms of data analysis.
Now that we've covered the basics of handling missing data, we're ready to move on. In the next tutorial, we'll talk about another method for smoothing out data that will let us retain our monthly data: Rolling statistics. This will be useful for smoothing out our data as well as gathering some basic statistics on it.
There exists 2 quiz/question(s) for this tutorial. for access to these, video downloads, and no ads.
-
Data Analysis with Python and Pandas Tutorial Introduction
-
Pandas Basics - p.2 Data Analysis with Python and Pandas Tutorial
-
IO Basics - p.3 Data Analysis with Python and Pandas Tutorial
-
Building dataset - p.4 Data Analysis with Python and Pandas Tutorial
-
Concatenating and Appending dataframes - p.5 Data Analysis with Python and Pandas Tutorial
-
Joining and Merging Dataframes - p.6 Data Analysis with Python and Pandas Tutorial
-
Pickling - p.7 Data Analysis with Python and Pandas Tutorial
-
Percent Change and Correlation Tables - p.8 Data Analysis with Python and Pandas Tutorial
-
Resampling - p.9 Data Analysis with Python and Pandas Tutorial
-
Handling Missing Data - p.10 Data Analysis with Python and Pandas Tutorial
-
Rolling statistics - p.11 Data Analysis with Python and Pandas Tutorial
-
Applying Comparison Operators to DataFrame - p.12 Data Analysis with Python and Pandas Tutorial
-
Joining 30 year mortgage rate - p.13 Data Analysis with Python and Pandas Tutorial
-
Adding other economic indicators - p.14 Data Analysis with Python and Pandas Tutorial
-
Rolling Apply and Mapping Functions - p.15 Data Analysis with Python and Pandas Tutorial
-
Scikit Learn Incorporation - p.16 Data Analysis with Python and Pandas Tutorial